Comparative Genomics: Analysis of Evolutionary Relationships Among Species
Written by Amélie Lagacé-O'Connor | Published 2023-8-19
Written by Amélie Lagacé-O'Connor | Published 2023-8-19
This blog post will explore the field of comparative genomics, which involves comparing the genomes of different species to understand their evolutionary relationships and the mechanisms of genetic adaptation. It will discuss some of the key tools and techniques used in comparative genomics, and the insights that have been gained from studying the genomes of diverse organisms.
Comparative genomics, a captivating domain within biological research, involves the comparison of genomes across diverse species. The analytical approach unfurls profound insights into evolutionary relationships, genetic variations, and functional elements. Functional elements, discrete sequences within a genome, orchestrate vital roles in an organism’s biological processes. The genome, encompassing an organism’s entirety of genetic material, includes both genes and non-coding regions, each holding a distinct piece of the puzzle.
Through this lens, scientists navigate the genetic landscapes to unveil shared traits and unique genetic differences, uncovering not only patterns in sequences but also the architectures of genes and genetic attributes. This information provides valuable insights spanning diverse fields—from tracing evolutionary bonds to deciphering gene regulation, exploring genetic variations and disease susceptibility, to unraveling the structural intricacies encoded in a genome. Moreover, these insights ripple into functional annotation and the intricate tapestry of phylogenetics.
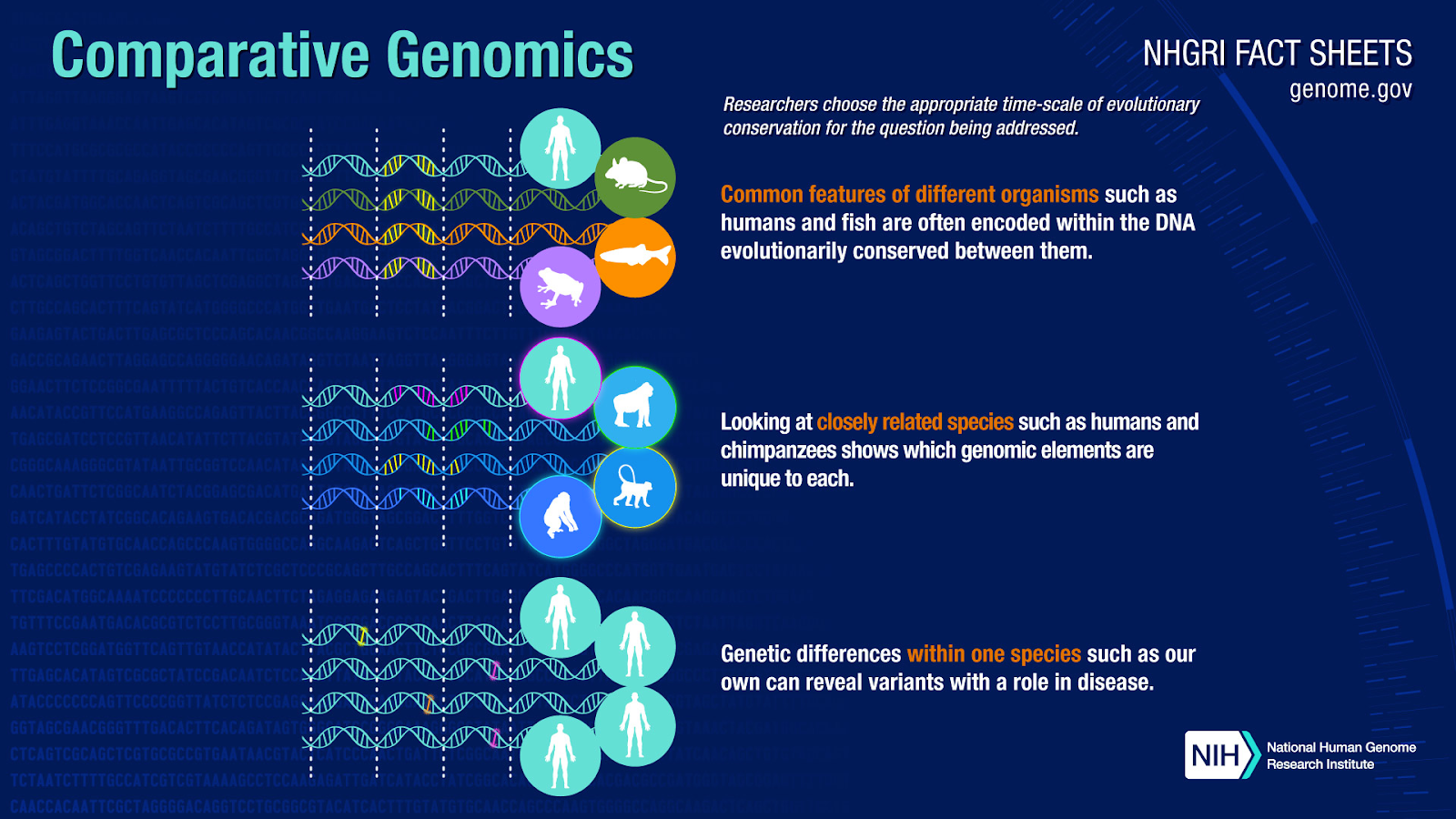
Insights from comparative genomics resonate profoundly in human welfare, translating genetic blueprints into practical applications. Bridging this knowledge to real-world impact, researchers link comparative genomics with drug discovery and personalized medicine, showcasing the transformation of insights into tangible change. This illustrates the significance of comparative genomics and the value of advancing the field’s technology.
Comparative genomic researchers initially employed the technique of sequence alignment, comparing the DNA or protein of distinct organisms through sequence alignment algorithms to identify the regions of similarity and difference. In the absence of computers, researchers manually pinpointed these crucial regions. Armed with much precision, they unveiled patterns within sequences, shaping deeper understanding of the connections between organisms. Nonetheless, this labor-intensive process was very time-consuming and limited in managing extensive datasets.
Fast-forward to the landscape of contemporary comparative genomics, there is a lot of reliance on high-throughput sequencing technologies, particularly in next-generation sequencing (NGS) and more recently, third-generation sequencing (long-read sequencing). NGS encompasses techniques enabling the simultaneous sequencing of thousands to millions of DNA fragments, generating substantial sequence data swiftly and cost-effectively. Prominent NGS platforms include Illumina, Ion Torrent (by Thermo Fisher), and 454 (by Roche). This methodology entails sequential stages of library preparation, cluster generation, sequencing, and subsequent analysis. In contrast, third-generation sequencing (TGS) technologies were developed to analyze longer DNA fragments directly. The principles underlying TGS are upheld by using technologies like Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). This transformative approach starts with template preparation, which is attaching DNA samples to polymerase enzymes. Subsequently, it continues with sequencing, as DNA fragments pass through nanopores, while recording the nucleotide, ultimately culminating in data analysis.
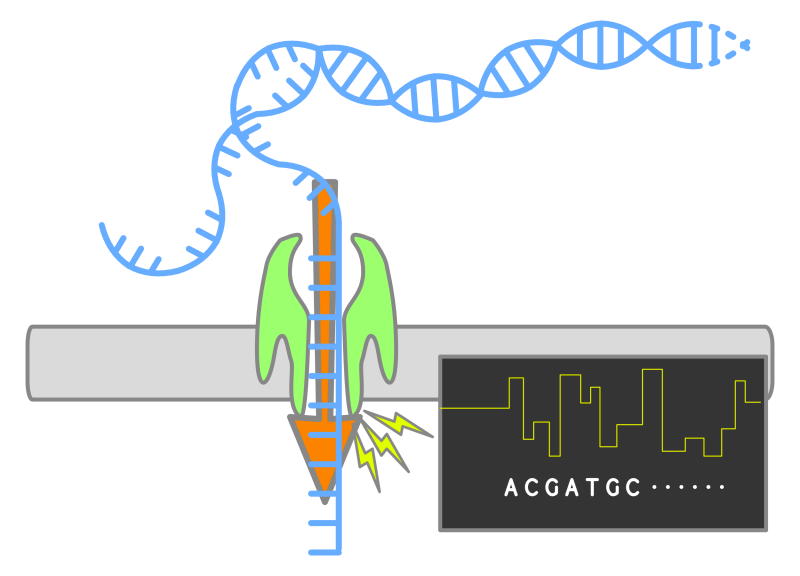
Next-generation sequencing boasts high throughput and accuracy. The extensive throughput facilitates simultaneous sequencing of large genomes and multiple samples, bolstering the research efficiency. Moreover, the precision and minimal errors of NGS instills confidence in analyses, enhancing result reliability. On the other hand, third-generation sequencing offers advantages of extended read lengths, single-molecule sequencing, and real-time sequencing. The longer read lengths are beneficial for assembling complex genomes, resolving repetitive regions, and analyzing structural variations. Single-molecule sequencing eliminates the need for amplification and mitigates potential biases, while real-time sequencing enables rapid data acquisition and dynamic monitoring of molecular events.
Comparative genomics has proven to be an instrumental tool, giving researchers invaluable insights spanning various domains. From deciphering the evolution of brain size and unraveling the mysteries of neurodevelopmental disorders to understanding the intricate propagation of viruses, notably the Ebola virus, this dynamic field stands as a beacon of discovery. Evidencing its magnitude, the 2014 Ebola outbreak in West Africa holds the record as the largest documented outbreak for this virus, as outlined in a CDC article. In the battle against this pathogenic threat, researchers turned to comparative genomics to navigate the intricacies of the proteins involved in their mechanisms.
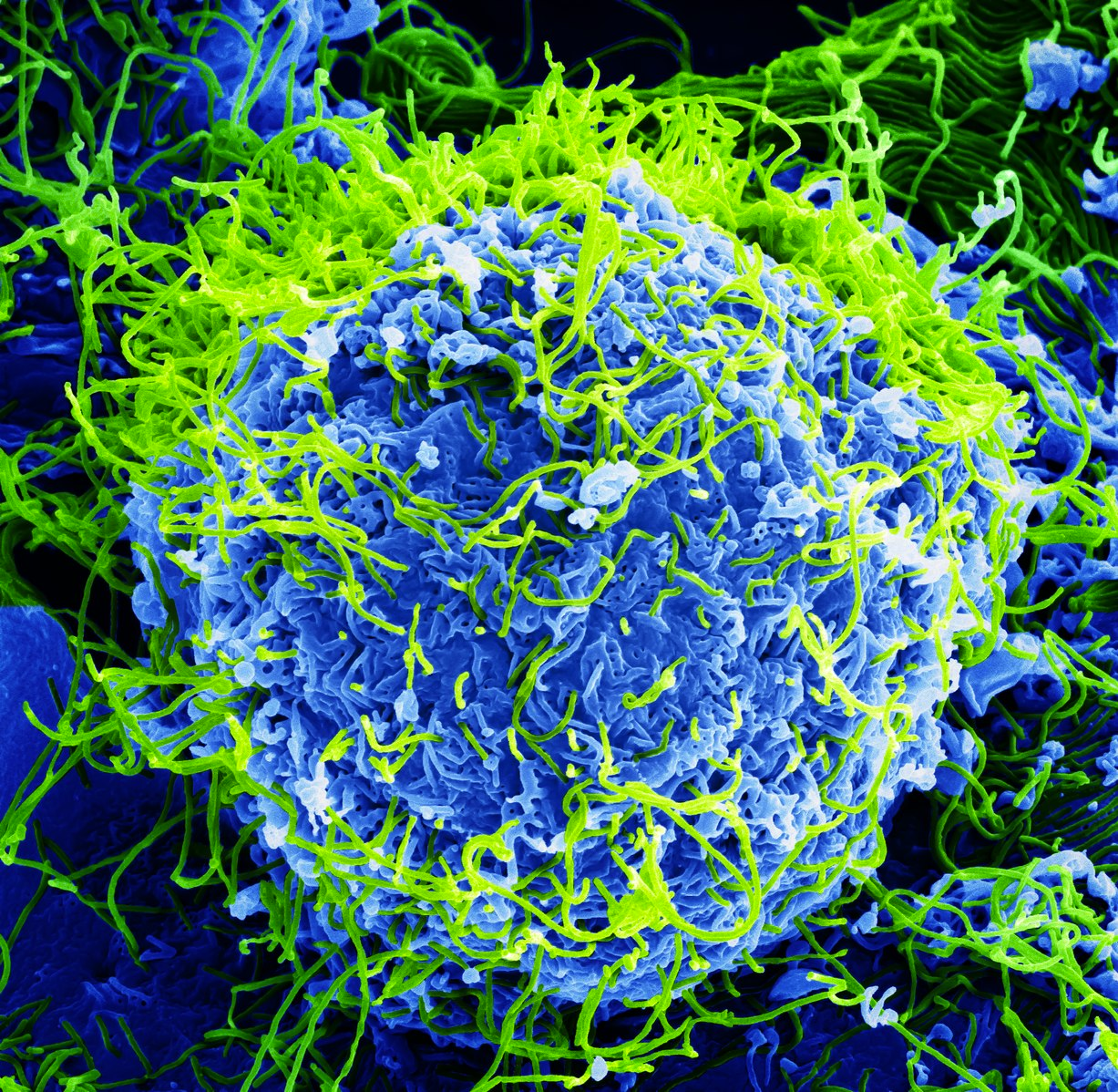
In this pursuit, researchers evaluated over 100 available ebolavirus virus genomes, comparing them with one another and to other viruses. Utilizing oligomer frequency analysis, a computational technique applied following tools like next-generation sequencing or third-generation sequencing, they found the Filoviridae virus family as a distinctive cluster including Ebolavirus, as described in a 2015 article by Jun et al. The analysis also uncovered genomes within this family encoding protein of similar functions and gene arrangement. Amidst these revelations, differences surfaced in specific genome regions of the virus, such as those encoding glycoprotein, nucleoprotein, and polymerase. These variations painted a roadmap, guiding researchers towards the belief that these regions could be potential therapeutic targets. This example highlights the pivotal role of comparative genomics, not only in crafting effective therapeutics but also in shaping numerous other fields.
The realm of comparative genomics emerges as a remarkable force driving scientific exploration. We have seen it used to decipher genetic intricacies, unraveling evolutionary mysteries, decoding the mechanisms behind neurodevelopmental disorders, and even shining a light on the propagation dynamics of viruses like the Ebola virus.In the 2014 outbreak of this pathogen, guided by the technology of oligomer frequency analysis, researchers were able to find exploitable areas that could be used to develop therapeutics against such a dangerous virus, demonstrating the importance of comparative genomics.
By Danial Gharaie Amirabadi
By Colorado Wilson
By Keaun Amani
By Amélie Lagacé-O'Connor
By Keaun Amani
By Danial Gharaie Amirabadi
Register for free — upgrade anytime.
Interested in getting a license? Contact Sales.
Sign up free