Molecular Dynamics Experiments with GROMACS on the Neurosnap Platform
Written by Colorado Wilson | Published 2024-1-23
Written by Colorado Wilson | Published 2024-1-23
Molecular dynamic simulations have a deep history dating back to techniques and equations of motion developed by Isaac Newton, who was interested in N-body systems. Many of these techniques and methods precede the advent of modern computing. Many simulations and developments laid the foundation for work that would begin in the 1970s, using molecular dynamic simulations in biochemistry. Simulations quickly grew from hundreds of atoms to complete protein structures with millions of atoms. Now, simulations can be done across many different software, such as GROMACS, AMBER, NAMD, CHARMM, LAMMPS, and many more, with millions of atoms. The expansion of available software and possible simulations led to the explosion of molecular dynamics applications. Molecular dynamics has uses for a broad array of research tasks, including drug discovery, understanding biomolecule structures and kinetics, complex interactions, and more. Here on the Neurosnap platform, we deploy GROMACS for biochemical simulations and bear the burden of setting up complex software environments.
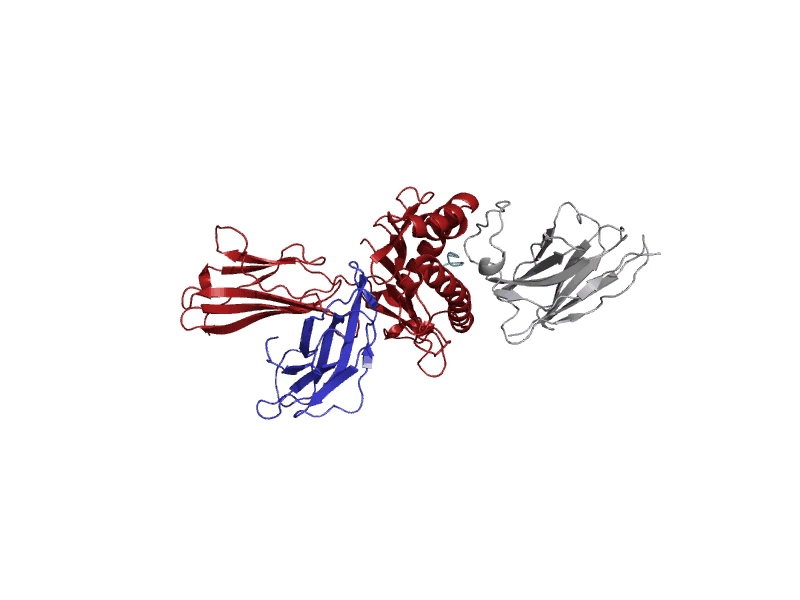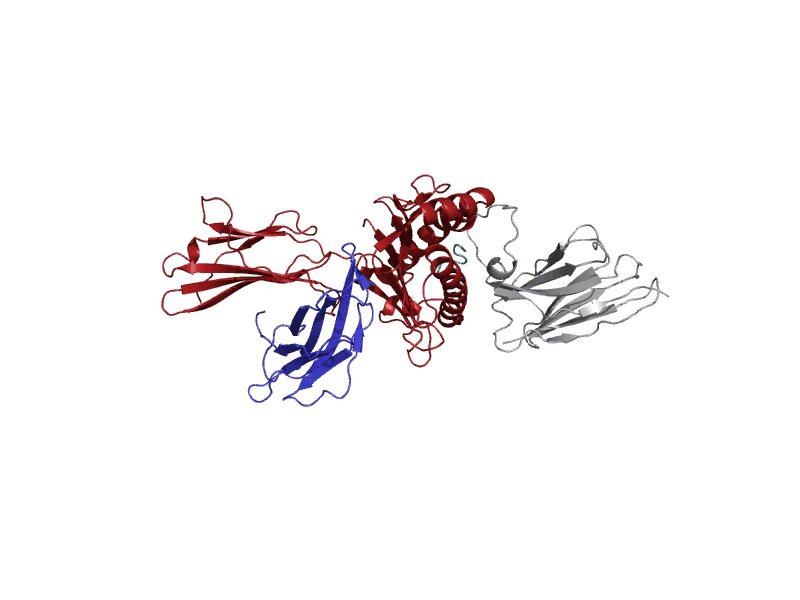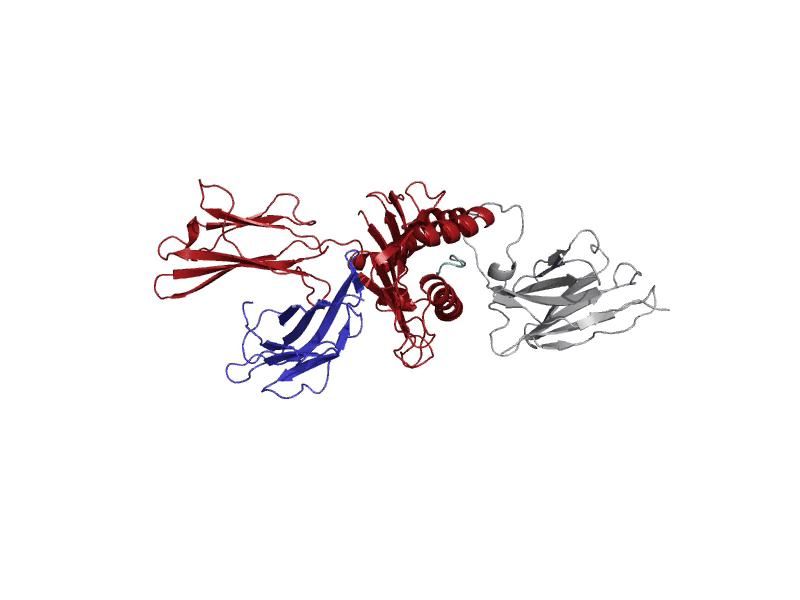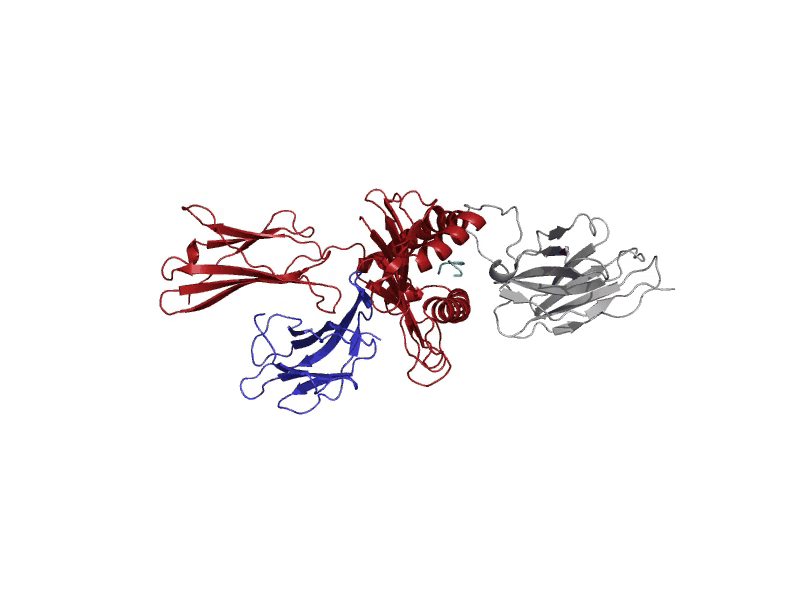
GROMACS allows for diverse molecular dynamic experiments under realistic temperature, pressure, and solvent conditions. These conditions may resemble biological states or other more complex environments.
GROMACS is a C-based program that can be GPU-accelerated and is operated via the command line. The GROMACS software was developed in 1991 by the Department of Biophysical Chemistry at the University of Groningen in the Netherlands. As of 2001, GROMACS is managed and updated by Berk Hess at the Royal Institute of Technology and Erik Lindahl at Stockholm University.
GROMACS via the Neurosnap platform is available to Premium subscribers as it is compute and time extensive. GROMACS can be found on our services page along with all the other models; click on the tile, and you will land on the input page. The input page is where the molecular dynamics experiment is designed, starting with an input PDB file containing the structure of interest. If the experimental design is focused on a protein-ligand complex, the Neurosnap platform will tailor the experiment to dial in on that interaction. Lastly, the input page allows control over the solvent shape, charge, and concentration. After designing the experiment, Neurosnap will take care of the rest.
GROMACS produces an immense amount of data during its simulation, and digesting it can be complicated. The Neurosnap platform uses a handful of simple figures to display the most meaningful data produced throughout the 1000 picosecond (1 nanosecond) experiment. The Neurosnap figures can be broken into three segments: structures, equilibration data, and simulation data.
Structures:
A molecular dynamics run will log "frames" of the simulation that capture the structure at set intervals. These frames are stored in the trajectory file, "md_center.xtc", returned to the user after the experiment. Each of these frames serves to understand where the atoms are throughout the experiment. After the experiment, the last frame after 1000 picoseconds will be displayed in the interactive protein viewer and contains all the atoms, including solvent and ions. It is recommended for quick viewing to hide the solvent and ion atoms. This final frame is also returned to the user as a PDB file, "output.pdb".
Simulation Data:
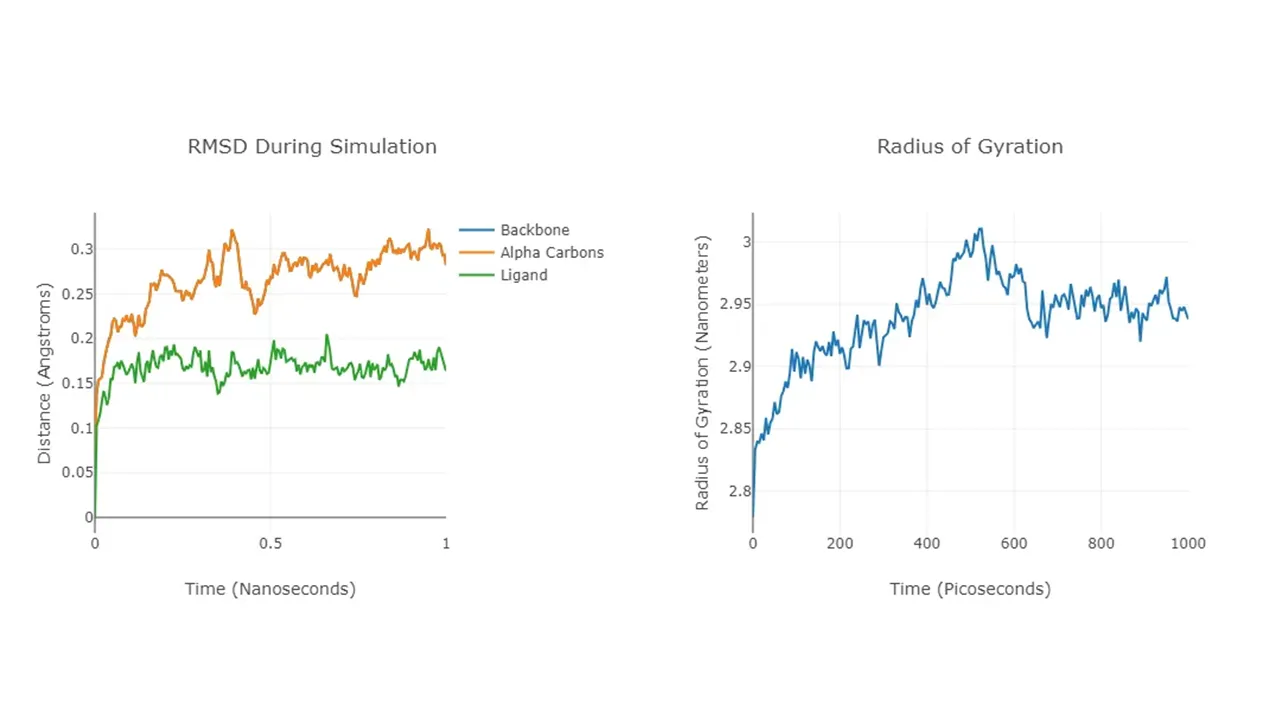
During the simulation, the RMSD of the entire complex is recorded, and if it's a protein-ligand experiment, the ligand is also recorded individually. This metric is, at its core, a measurement of stability. A high RMSD or high variation in RMSD indicates instability in the conformation of the structure and signals significant changes throughout the simulation. The radius of gyration (Rg) is calculated over the run and is another stability measurement; spikes in Rg indicate an unfolding event, while low variance indicates a stable fold.
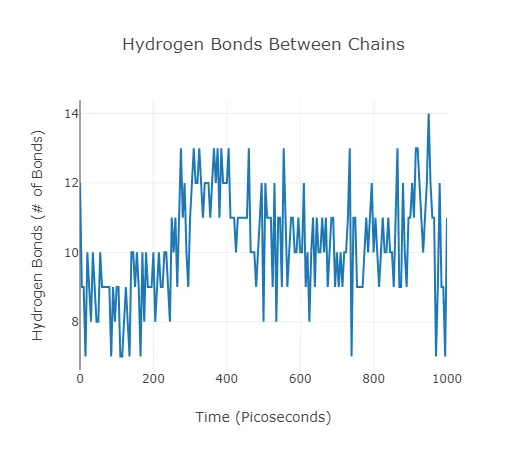
As mentioned, when an experiment is marked as a protein-ligand simulation, Neurosnap orients the experiment around the chains of interest for the binding interaction. The two chains containing the ligand and protein binding interface will have their hydrogen bonds recorded and plotted over the simulation. These Hydrogen Bonds are an indicator of the thermodynamic stability of the complex.
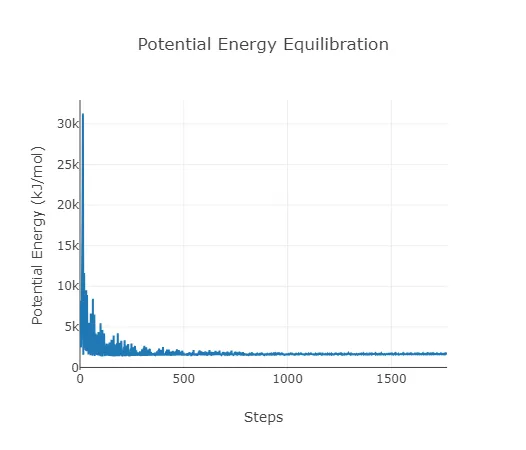
Equilibration Data:
During the simulation, several equilibration steps take place to find the target temperature, pressure, and potential energy for the simulation itself. The temperature and pressure are stabilized through thermostat and barostat algorithms using NVT and NPT ensemble methods. The potential energy is stabilized by translating atoms to less conflicting positions, reducing clashes, and producing a more physically stable structure. The temperature, pressure, and potential energy should roughly converge to a stable measurement, and if they don't, it's a sign that your structure or experimental conditions are not stable.
GROMACS on the Neurosnap platform combines our intuitive interface with the high complexity of GROMACS to allow researchers to perform molecular dynamic experiments. Neurosnap takes the compute and time burden off the researcher and accelerates their work, resulting in easy-to-digest information about the equilibration, simulation, and analysis of GROMACS molecular dynamics experiments.
By Keaun Amani
By Amélie Lagacé-O'Connor
By Danial Gharaie Amirabadi
By Danial Gharaie Amirabadi
By Danial Gharaie Amirabadi
By Danial Gharaie Amirabadi
Register for free — upgrade anytime.
Interested in getting a license? Contact Sales.
Sign up free