What Is Inverse Folding & How To Practically Apply It
Written by Keaun Amani | Published 2023-12-30
Written by Keaun Amani | Published 2023-12-30
Discover the cutting-edge realm of inverse folding models—an innovative and potent tool transforming the de-novo protein design landscape. These models have seamlessly contributed to the creation of therapeutic agents, biosensors, and industrial enzymes. Join us in this blog post as we delve into the comprehensive understanding of inverse folding, exploring its applications and unveiling the optimal utilization of inverse folding models on Neurosnap for unparalleled precision in protein design.
Let's start off by talking about Protein Folding. If you're not familiar with the concept, protein folding is the process wherein a polypeptide or protein gracefully transforms into a three-dimensional conformational state, commonly referred to as the protein's structure. Advanced techniques, such as AlphaFold2 and other cutting-edge protein structure prediction models, play a pivotal role in this domain. These models operate by taking the amino acid (AA) sequence of a given protein as input and employing sophisticated algorithms to predict their intricate structure or fold. It's a fascinating journey into the molecular intricacies that govern the formation of these essential biological building blocks.

As the name entails, inverse folding does the exact opposite of protein folding by trying to predict a sequence that could fold into a desired structure. Typically these models are trained on massive datasets of masked protein structures that the models then have to predict the original sequences of. Masking a protein structure is the process of hiding all the side chains found within structure so that only the backbone of the protein remains. This typically means that the only remaining atoms are the alpha-carbon, beta-carbon, and the nitrogens that are essential to the backbone and nothing else. Additionally, some models like ProteinMPNN also add some noise or random variance to the coordinates of these atoms to make the model more robust and resistant to overfitting. This masked protein or protein backbone is also the input the model receives during inference or prediction stages.
While this might seem counterintuitive this is actually an incredibly useful process for protein design, especially for enzymes and therapeutics. For starters, inverse folding models tend to be very fast and can predict hundreds of sequences that might fold into a desired structure within mere minutes. Additionally, the sequence identities between resulting proteins is usually between >0.4 and <0.75 which is very valuable for enzyme design as it allows you to sample a much broader portion of the sequence space compared to traditional methods that might rely on only a few point mutations or indels.
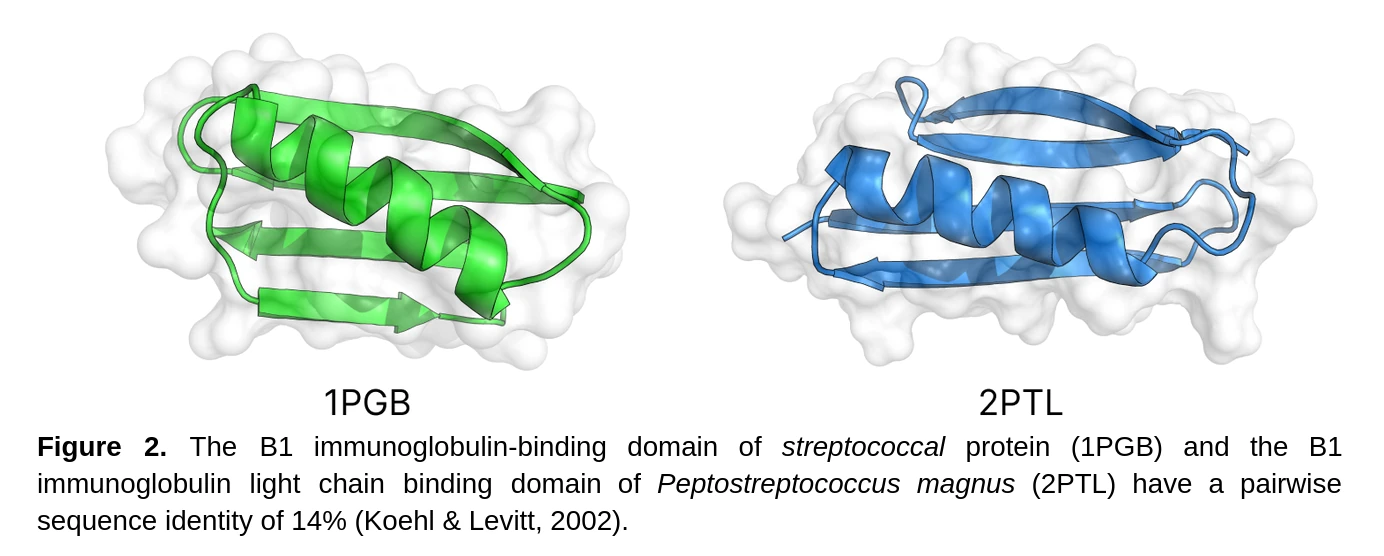
Additionally, models like ProteinMPNN are autoregressive and work well with complexes allowing you to fix specific chains which is valuable for designing therapeutics such as peptides and small protein binders. For example you could use tools like RFdiffusion to design peptides and mini-binders that interact with a specific region of a target protein or receptor. After this an inverse folding model can be used to create sequences that will produce the desired complex which can then be screened for in follow up steps.
Another benefit of some inverse folding models like ProteinMPNN and ESM-IF1 is that they come with confidence metrics that can be used gauge how reliable a prediction is.
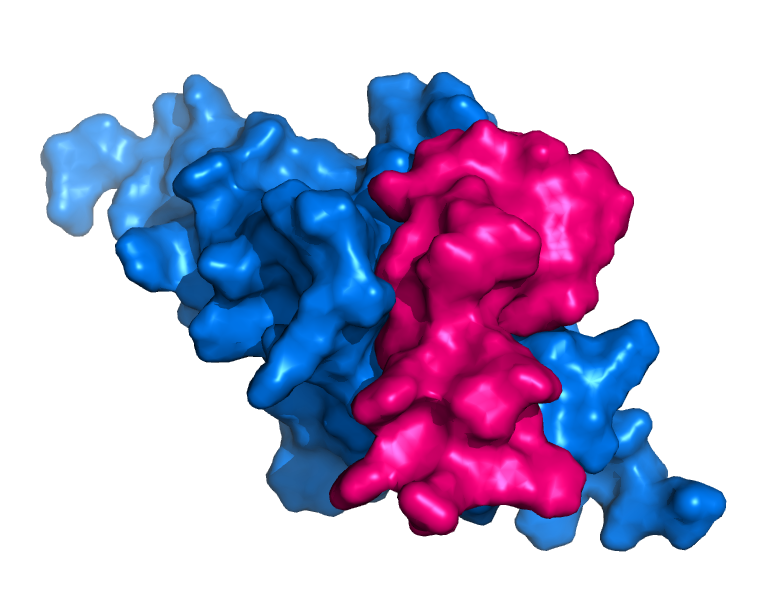
Now that we've covered the basics of what inverse folding is, lets dig into some tips and tricks on how to use these models effectively for our research goals.
Admittedly, ProteinMPNN has been found to struggle with certain proteins and complexes, producing non-sense output sequences such as those with many repeats or residues like cysteines in places where there really shouldn't be any cysteines. A technique that can be applied to fix a lot of these issues is by increasing the number of amino acids that are visible to the model. Remember that all worthwhile inverse folding models essentially mask the input structure before trying to predict corresponding sequences. For autoregressive models like ProteinMPNN, you can actually specify which amino acids can be "fixed" or fed into the model. Fixed positions are amino acids that are not masked and are fed directly into the model to bias its outputs.
Depending on your use case, fixing specific domains, chains, or simply just fixing a random percentage of positions within the structure can bias ProteinMPNN just enough to fix the issue. Another useful trick is to fix positions that form loops and other flexible parts of the protein as ProteinMPNN has been observed to sometimes place rigid amino acids like Histidine in those positions or other disruptive amino acids such as Tryptophan or Phenylalanine.
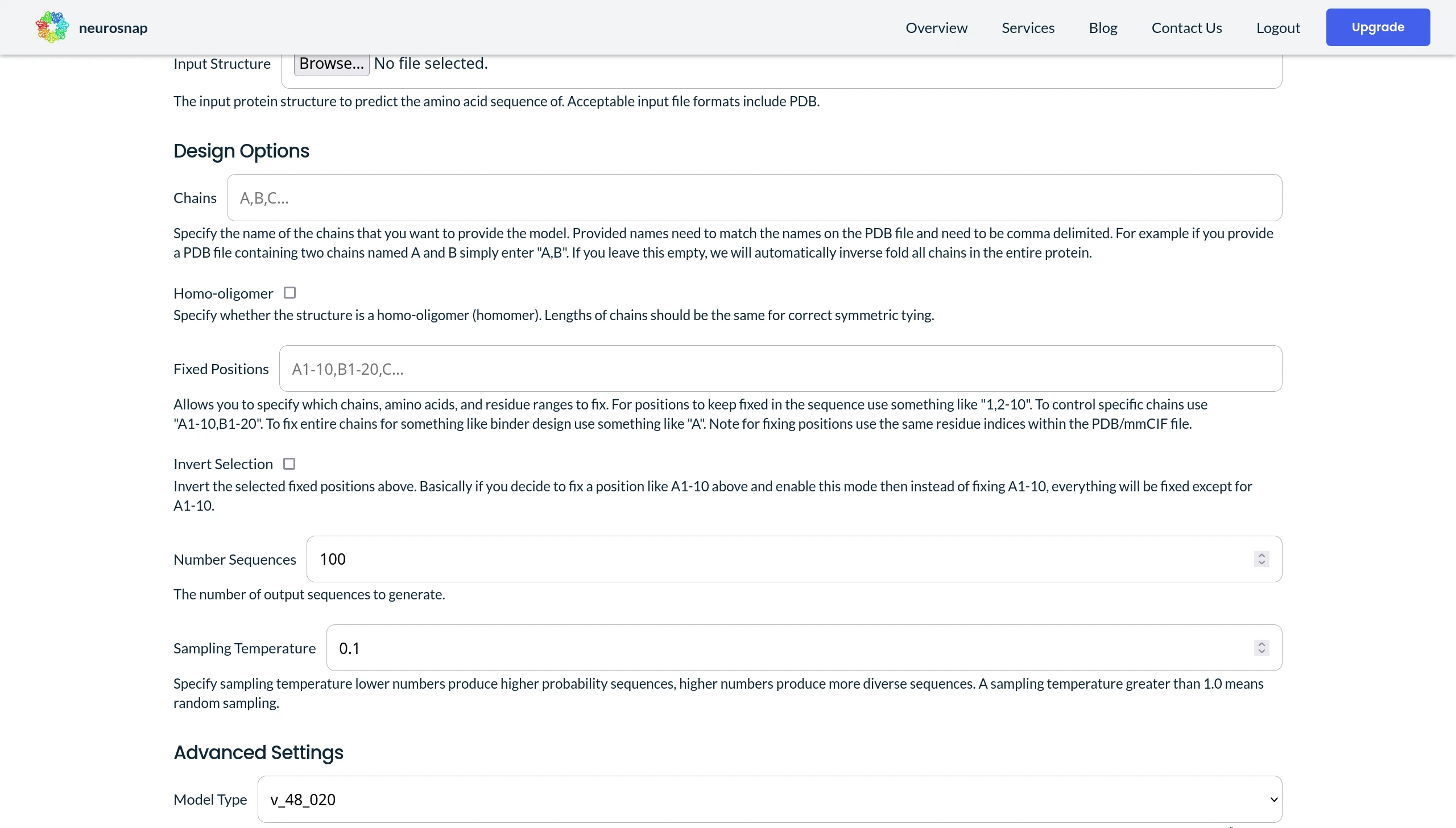
One practical option to control for cysteines or any other problematic amino acids is to simply bias the model to remove them from all predictions. On Neurosnap this can easily be done by specifying C in the Excluded Amino Acids field for the ProteinMPNN input panel.
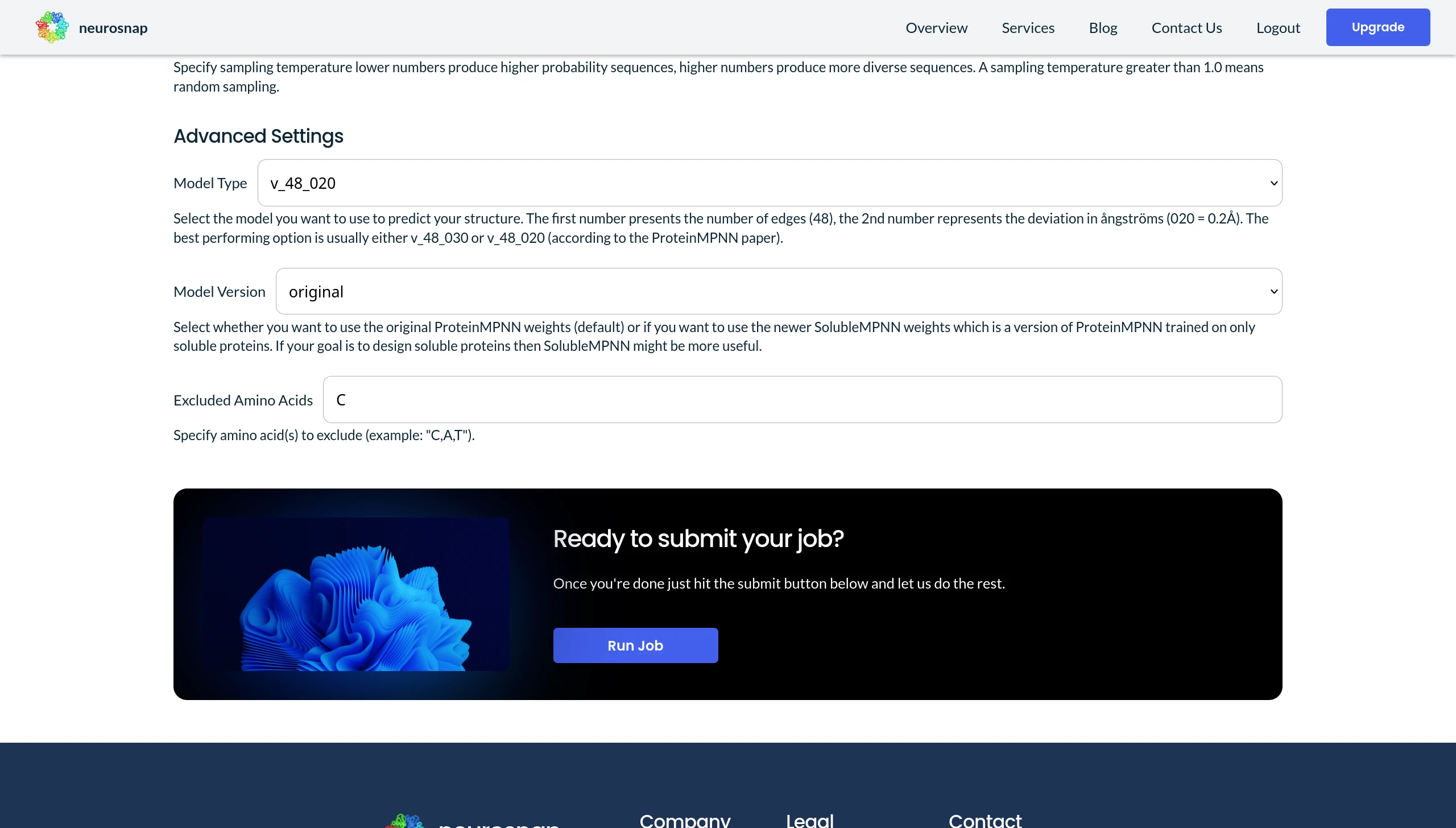
An established practice involves optimizing the solubility of enzymes or proteins. Interestingly, this can also be accomplished using ProteinMPNN. A specialized iteration of ProteinMPNN, explicitly trained on soluble proteins, proves invaluable for generating variants of a target protein/enzyme. This tailored model excels at predicting proteins exhibiting similar structures and heightened solubility. To leverage this capability on Neurosnap, simply switch the 'Model Version' to 'soluble'.
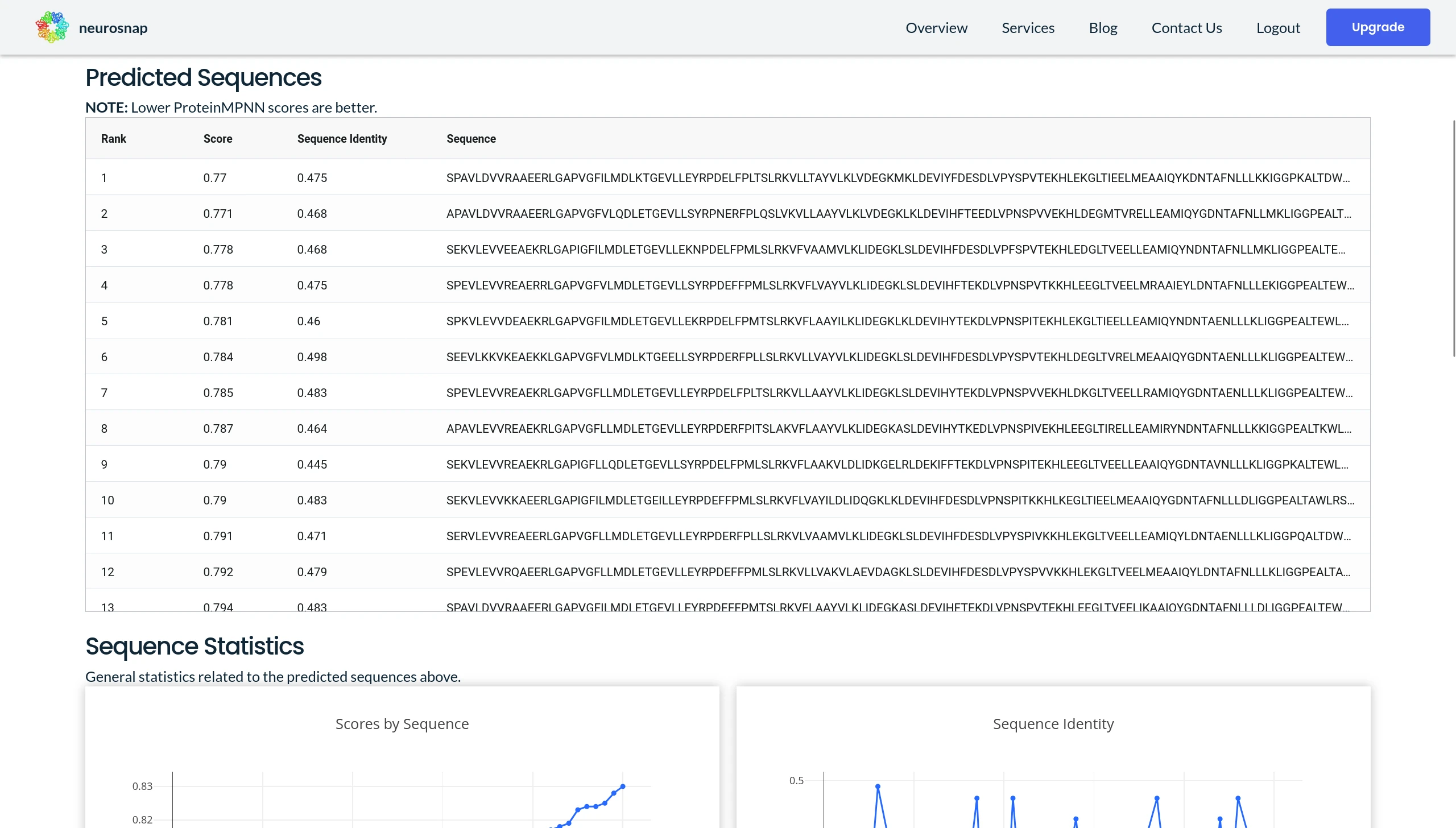
One of the best ways to evaluate ProteinMPNN predictions is to filter sequences by their Score. The Score is an output metric from ProteinMPNN where values closer to 0 tend to result in better predictions. This is generally a good start for filtering candidates. After you've selected the top x number of sequences from ProteinMPNN, a good follow up step is to then predict their structures with AlphaFold2 and then measure the TM-Align score between the original protein and the variants. Generally speaking you want your proteins to possess similar structures as proteins with similar structures tend to behave similarly. Also note that AlphaFold2 isn't designed for predicting the effects of mutations and in some cases might produce misleading results. This is why we recommend also performing experiments to validate any hypotheses.
Now that you've learned how to effectively use ProteinMPNN as well as the applications of inverse folding models, we suggest reading our blog post on applying the concepts above for producing variants of Aausfp1.
Want to get started with running ProteinMPNN or AlphaFold2? Register here and run your own jobs!
By Keaun Amani
By Keaun Amani
By Danial Gharaie Amirabadi
By Danial Gharaie Amirabadi
By Danial Gharaie Amirabadi
By Keaun Amani
Register for free — upgrade anytime.
Interested in getting a license? Contact Sales.
Sign up free