Introducing Chai-1: A Commercial Alternative to AlphaFold3
Written by Danial Gharaie Amirabadi | Published 2024-11-28
Written by Danial Gharaie Amirabadi | Published 2024-11-28
The field of molecular structure prediction has seen remarkable progress in recent years, and a significant player in this space is Chai-1. Chai-1 is a multi-modal foundation model designed for biomolecular structure prediction, excelling in tasks such as predicting protein-ligand and protein multimer interactions. Unlike AlphaFold3, which has restrictions on its commercial use, Chai-1 is available as an open model, making it suitable for a wide range of applications, including commercial research projects. Here, we explore what makes Chai-1 a compelling tool and how Neurosnap can help you leverage it for your specific needs.
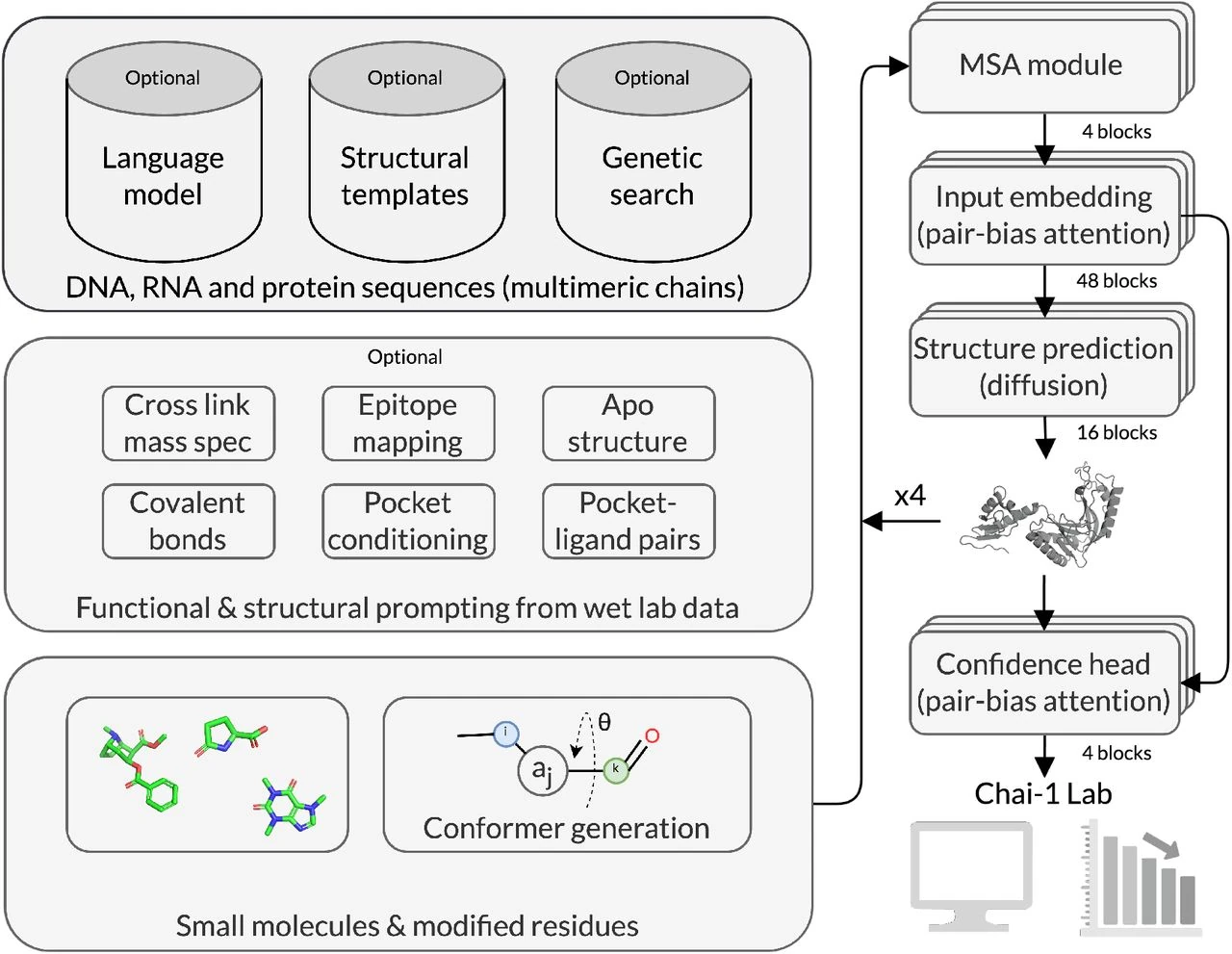
Chai-1 is a state-of-the-art model that predicts the structure of biomolecules, such as proteins and nucleic acids, using both raw sequence and chemical input data. It is designed to be versatile and high-performing across several structure prediction tasks, which are crucial for drug discovery and understanding cellular mechanisms. Notably, Chai-1 supports single-sequence prediction without relying on multiple sequence alignments (MSAs), allowing it to produce strong predictions even with minimal evolutionary information, outperforming models like ESMFold in these conditions.
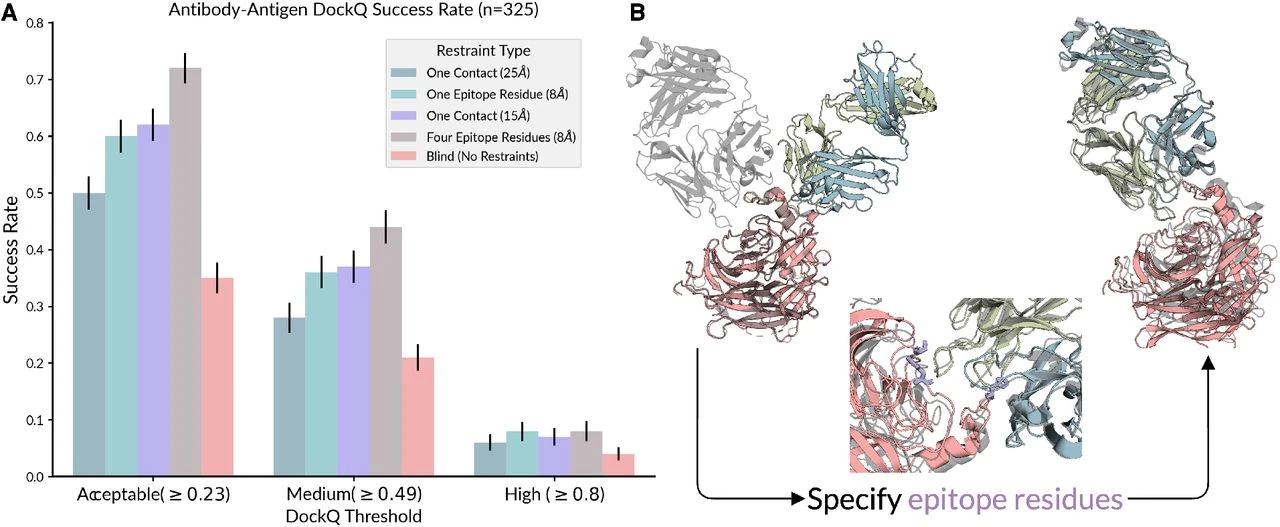
Chai-1 can also be optionally prompted with experimental constraints, such as data from cross-linking mass spectrometry or epitope mapping, to further improve its prediction accuracy in complex scenarios. This added flexibility makes Chai-1 an ideal tool for researchers looking for enhanced structure prediction capabilities without the cumbersome licensing limitations associated with some of its competitors.
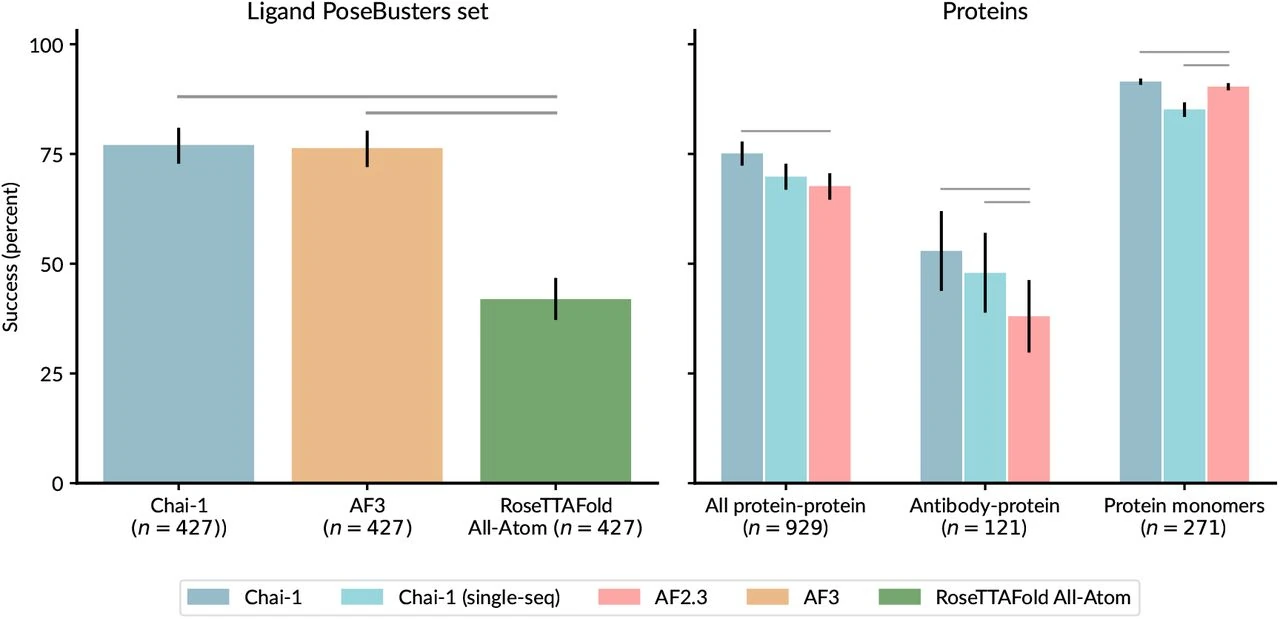
Chai-1 competes directly with AlphaFold3 in terms of prediction accuracy, particularly for protein-ligand interactions and multimeric protein complexes. For example, in evaluating ligand binding predictions, Chai-1 has shown success rates comparable to AlphaFold3 while providing the benefit of fewer restrictions on usage. Where AlphaFold3 might limit commercial implementation, Chai-1 allows for a more open approach, making it a more suitable option for research organizations seeking flexibility in their commercial applications.
| Method | Protein-protein | Protein-antibody | Protein monomers |
|---|---|---|---|
| Chai-1 (with MSA) | 0.751 (0.723, 0.778) | 0.529 (0.438, 0.620) | 0.915 (0.907, 0.922) |
| Chai-1 (single-sequence) | 0.698 (0.668, 0.728) | 0.479 (0.388, 0.570) | 0.852 (0.834, 0.867) |
| AlphaFold 2.3 multimer | 0.677 (0.646, 0.706) | 0.380 (0.298, 0.463) | 0.903 (0.895, 0.911) |
Additionally, Chai-1's ability to work in single-sequence mode, without the need for MSAs, allows it to provide faster and comparably accurate predictions even for variable or novel protein sequences. This can be particularly advantageous in contexts where traditional evolutionary information is sparse or unavailable.
At Neurosnap, we recognize the potential of Chai-1 to transform the biomolecular modeling landscape, particularly for commercial research. We are pleased to offer Chai-1 as part of our services, enabling companies and research teams to utilize this advanced model for their projects in drug discovery, protein engineering, and beyond.
By working with Neurosnap, clients can access Chai-1 through an intuitive and user-friendly service without needing to worry about implementation details or infrastructure. This allows you to focus on scientific insights and discoveries, while we handle the complexity of running and maintaining the tool.
Access our Chai-1 using our easy to use web UI.
By Keaun Amani
By Keaun Amani
By Danial Gharaie Amirabadi
By Keaun Amani
By Amélie Lagacé-O'Connor
By Danial Gharaie Amirabadi
Register for free — upgrade anytime.
Interested in getting a license? Contact Sales.
Sign up free