Achieving AlphaFold3-Level Accuracy with Open-Source Boltz-1
Written by Danial Gharaie Amirabadi | Published 2024-11-21
Written by Danial Gharaie Amirabadi | Published 2024-11-21
Accurate models of biomolecular complexes are critical for understanding cellular functions and advancing therapeutics. While AlphaFold2 revolutionized protein structure prediction, the ability to accurately predict diverse biomolecular assemblies—such as those involving ligands, ions, and nucleic acids—remains a significant challenge. Existing methods either lack the robustness of physics-based approaches or are too narrowly specialized, limiting their utility for diverse biomolecular structures.
The introduction of AlphaFold3 marked a leap forward by achieving high accuracy across nearly all molecular types in the Protein Data Bank (PDB). However, its restrictive non-commercial license has left a gap for open, accessible tools that rival its capabilities.
Enter Boltz-1, a fully open-source, commercially accessible model that achieves AlphaFold3-level accuracy. Boltz-1 democratizes access to advanced biomolecular modeling, making it a powerful tool for researchers and industries alike.
Boltz-1 brings significant algorithmic innovations that enable it to achieve accuracy on par with AlphaFold3 while offering enhanced flexibility and computational efficiency. These advancements make it an invaluable resource for predicting complex biomolecular interactions.
These innovations allow Boltz-1 to deliver state-of-the-art accuracy while reducing computational overhead, making it both accessible and scalable.
Boltz-1 has demonstrated exceptional performance across diverse biomolecular benchmarks, rivaling proprietary tools like Chai-1, a non-commercial replication of AlphaFold3.
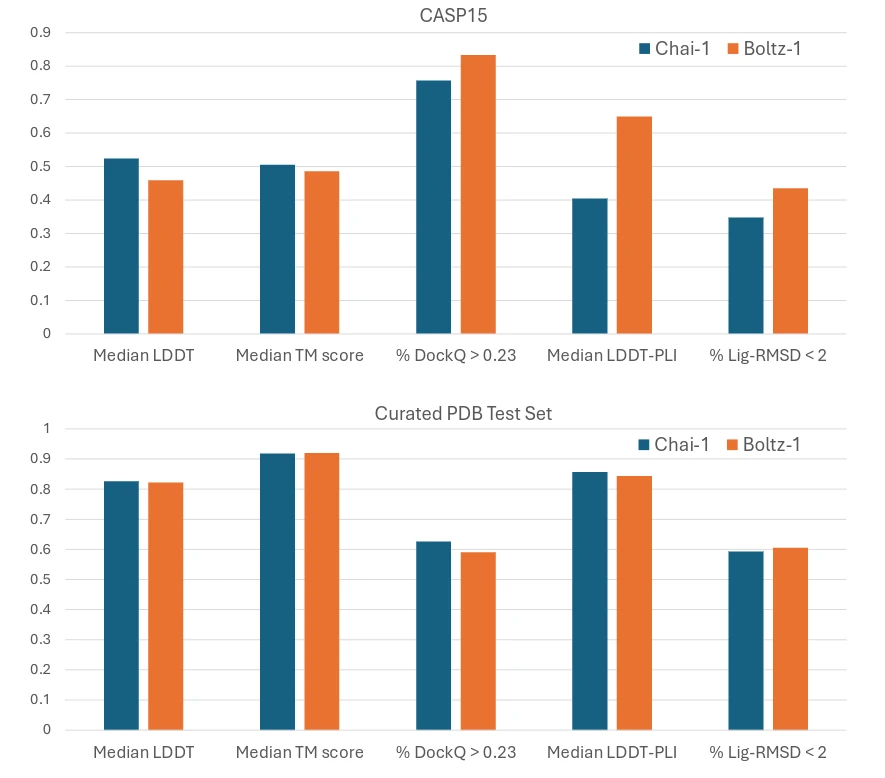
Boltz-1’s open-source framework and MIT license empower researchers, developers, and industries to explore and innovate without restrictions. Its performance and accessibility make it an ideal tool for advancing fields such as drug discovery, structural biology, and protein design.
Moreover, its ability to handle diverse biomolecular interactions makes it a versatile choice for academic and commercial applications alike.
Boltz-1 is now available via Neurosnap. Access AlphaFold3-level accuracy with ease. Learn more: Neurosnap Boltz-1 Service
By Danial Gharaie Amirabadi
By Amélie Lagacé-O'Connor
By Amélie Lagacé-O'Connor
By Danial Gharaie Amirabadi
By Keaun Amani
By Keaun Amani
Register for free — upgrade anytime.
Interested in getting a license? Contact Sales.
Sign up free